Evaluation of embryos in vitro has significantly improved over the past years with the introduction of time-lapse technology, which provides us with new tools to assess the morphological characteristics and implantation potential of an embryo. This technology has generated comprehensive information on the different events occurring between static observation periods and how the timing of these events help predict the embryo’s viability to implant. The assessment of the embryo’s dynamic morphology and its specific timing has lead to the new term “morphokinetics,” which is a different way of grading embryos as compared to static scoring.
Morphokinetics versus Conventional Scoring
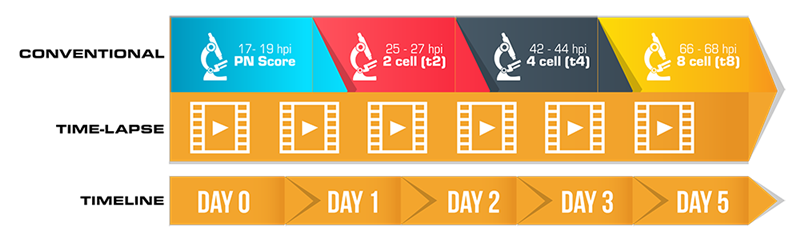
The conventional scoring system does not adequately cover what can be assessed with time-lapse. Time-lapse technology entails new assessment measures that allow for more precise grading due to dynamic monitoring and hence a more sophisticated way of grading embryos. This technology brought us new terminologies and assessment techniques that can’t be graded through conventional methods.
This article presents some basic terminologies that can be used by time-lapse users to help them achieve the full benefit of the technology. Using the same nomenclature across clinics will also allow us to compare and discuss data that result from annotations, scoring, and evaluation.
Morphokinetic Terminologies
-
Start of Time-Lapse (t0)
For conventional IVF, it is the time at which insemination occurs. For ICSI, it is the mid-time point from when injection begins and ends for that patient’s cohort of oocytes. Before you start the time-lapse, the MIRI® TL software will prompt you to choose whether it’s an ICSI or an IVF. This time point is important as the subsequent event timings are measured in hours post insemination/ injection, unless otherwise stated.
-
Pronuclear Fading (tPNf)
The time when both (or the last) PN disappear. This annotation is made at the first frame whereby the embryo is still at the 1-cell stage, but pronuclei can no longer be visualized.
-
Cleavage Events from t2 to t8
The time of the first cell cleavage is t2, and t3 is time for three discrete cells. Eight discrete cells will be annotated as t8. Annotate on the first frame when you first observe complete separation of the blastomeres.
-
Morula (tM)
This is the stage of compacted morula; wherein compaction can be full or partial. The status and time of compaction play a role on how the next stage, blastocyst, will develop.
-
Time of starting blastulation (tSB)
This is the initial stage of blastulation and the presence of a cavity can now be seen as a silver.
-
Time of full blastocyst (tB
The blastocyst starts to expand, and this is the stage before zona pellucida becomes thin.
-
Time of expanding blastocyst (tEB)
Blastocyst expansion happens in this stage, and zona pellucida becomes thin.
-
Time of hatching blastocyst (tHB)
This is the hatching of the embryo and extrusion of the embryo from the zona pellucida
Watch the video below as an example of how the embryo was annotated. You can go through again with the terminologies above until you become comfortable doing the annotation. The time markings for each event can then be compared with published models such as Meseguer (2011)[1], Rubio (2014)[2], Basile (2015)[3] or Desai (2014)[4] or with your laboratory-derived model. An increasing number of studies suggest that developmental timings differ between embryos with full developmental potential versus those with no or limited potential[5].
Annotation of different Morphokinetic Variables Using the MIRI® TL
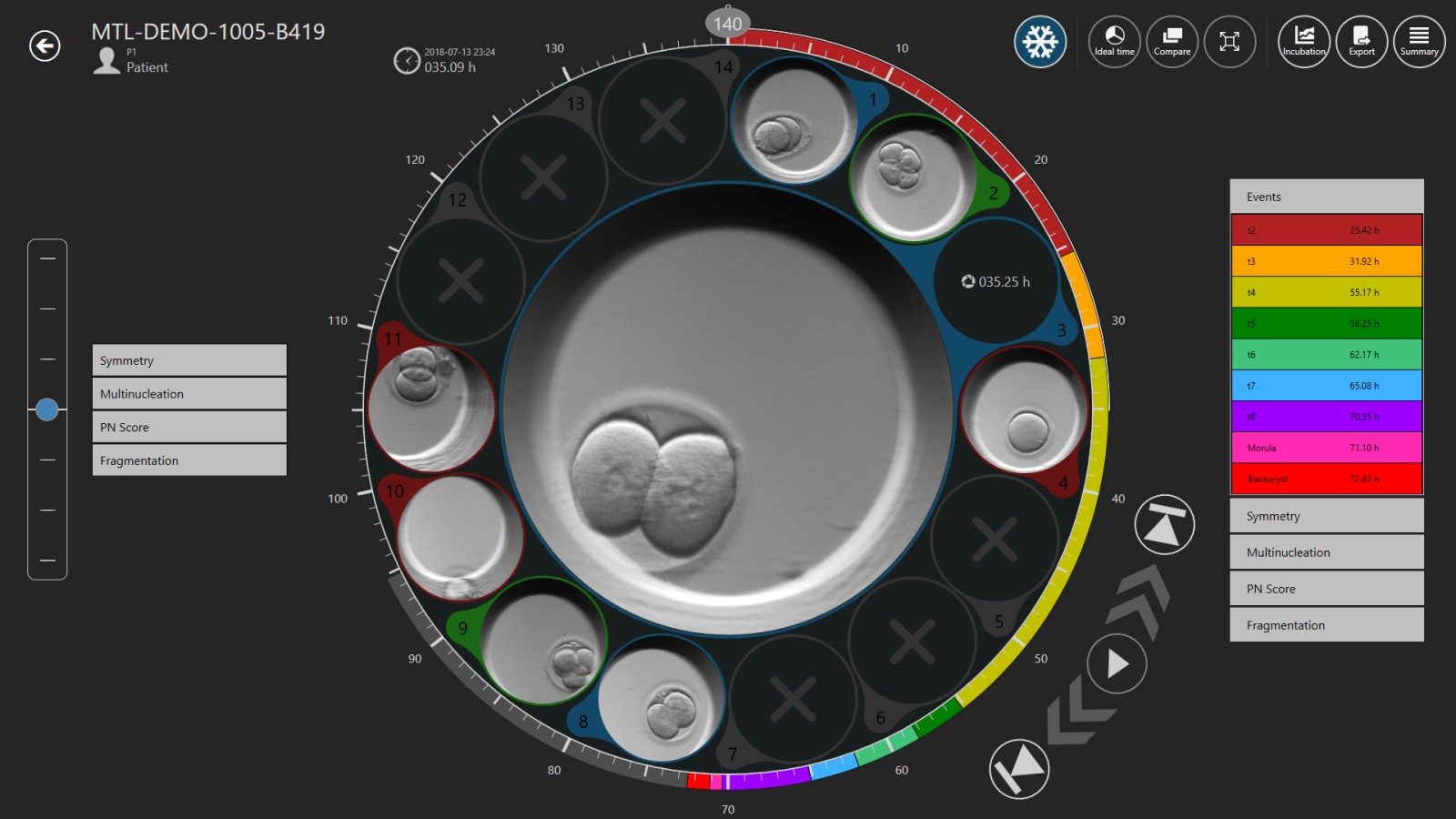
The MIRI® TL Viewer Software will display a set of events and parameters on the left-hand side, which you can use to annotate the embryo. Annotation is the process of time-marking a specific event/ parameter e.g. if you’ve observed t2 to happen at 27 hpi, you can click “t2” on the list of events from the left column and the annotated parameter of t2 at 27 hpi will be displayed in the right column. By default, the events list has t2, t3, t4, t5, 56, t7, t8, morula, blastocyst and early blastocyst. It is very important to accurately and consistently mark, annotate, these parameters as their values will be used by the software to automatically calculate length of cell cycles (cc) and synchrony (s) of cell divisions. If you feel you have made a mistake and wrongly annotated an event, simply double-click the annotation on the right hand side, it will return to the annotation list, on the left hand side, and you can re-annotate that event. Also, it is also very important that you annotate all events as the software cannot calculate cell-cycle lengths or synchrony of cell divisions without all events being annotated. Additional scoring tabs are also available for Symmetry, Multinucleation, PN Score and Fragmentation.
The MIRI® TL Viewer Software is adaptive. The events listed in the left column can be customized to include other parameters not included in the default settings. There are more advanced parameters that can be included in your time-lapse grading system. This article will only cover the events found in the MIRI® TL software default settings, which are practical and are commonly used by time-lapse users to have a better insight of the embryo’s development.
Summary of Morphokinetic variables and its definition (Source: Ciray et al., 2014[6])
|
Time |
Events |
|
t0 |
Time of IVF insemination or mid-time of micro/injection (ICSI/IMSI) |
|
tPNf |
Pronuclear fading. The embryo is at 1-cell stage and the appearance of pronuclei is no longer visible. |
|
t2 |
Time the cell divides into two cells. |
|
t3 |
3-cell stage. Marks the start of the second round cleavage. |
|
tM |
Morula stage, compaction process is complete. |
|
tSB |
Initial stage of blastulation, start of cavity formation. |
|
tB |
Full blastocyst, thinning of zona pellucida. |
|
tEB |
Expansion of the blastocyst, zona pellucida reduces to half. |
|
tHB |
Hatching of the blastocyst, embryo is now detached from zona pellucida. |
The Need for Consensus
Reliable selection of viable embryo plays a crucial role in the success of IVF treatment[5]. Using time-lapse technology, morphokinetic scoring aids in better selection of embryo. And with an increasing number of clinics using time-lapse technology, assessing the viability of embryo varies per clinic. A consensus scoring map to facilitate the assessment of embryo development is needed for consistency and better evaluation of embryo viability[2].
References
- Meseguer M, Herrero J, Tejera A, Hilligsoe K, Ramsing N, Remohi J (2011).
The use of morphokinetics as a predictor of embryo implantation. Human Reproduction, Vol. 26, No.10 pp. 2658-2771, 2011.
- Rubio I, Galan A, Larreategui Z, Ayerdi F, Bellver J, Herrero J, Meseguer M. (2014).
Clinical validation of embryo culture and selection by morphokinetic analysis: a randomized, controlled trial of the Embryoscope. Fertil Steril 2014; 102: 1287-1294.e5.
- Basile N, Vime P, Florensa M, Ruiz B, Velasco G, Remohi J, Meseguer M. (2015).
The use of morphokinetics as a predictor of implantation: a multicentric study to define and validate an algorithm for embryo selection. Human Reproduction, Vol. 30, No.2 pp. 276-283, 2015.
- Desai N, Ploskonka S, Goodman LR, Austin C, Goldberg J, Falcone T (2014).
Analysis of embryo morphokinetics, multinucleation and cleavage anomalies using continuous time-lapse monitoring in blastocyst transfer cycles. Reprod Biol Endocrinol 2014; 12: 54-9.
- Kirkegaard, K., Ahlstrom, A., Ingerslev, H., & Hardarson, T. (2014).
Choosing the best embryo by time lapse versus standard morphology. Elsevier Inc. http://dx.doi.org/10.1016/j.fertnstert.2014.11.003.
- Ciray, H., Campbell, A., Agerholm, I., Aguilar, J., Chamayou, S., Esbert, M., & Sayed, S. (2014).
Proposed guidelines on the nomenclature and annotation of dynamic human embryo monitoring by a time-lapse user group. Human Reproduction, Vol. 29, No.12 pp. 2650-2660, 2014.
